For antibody antigen modeling using the native version of HADDOCK3, I refer to the Antibody Antigen Modeling Using the Native Version of HADDOCK3 tutorial (Antibody-antigen modelling tutorial using a local version of HADDOCK3 – Bonvin Lab). Creating a HADDOCK3 profile for defining the docking workflow, no reference is given, how do I set up the caprieval step(s)?
Hi holly,
The [caprieval] module is mostly used for benchmarking purposes (if you have a reference file), or will use as reference the best scoring models.
Tt also allows to generate the final plots during the post-processing steps (in the analysis directory).
If you do not provide any reference file, just run [caprieval] using default parameters.
Cheers
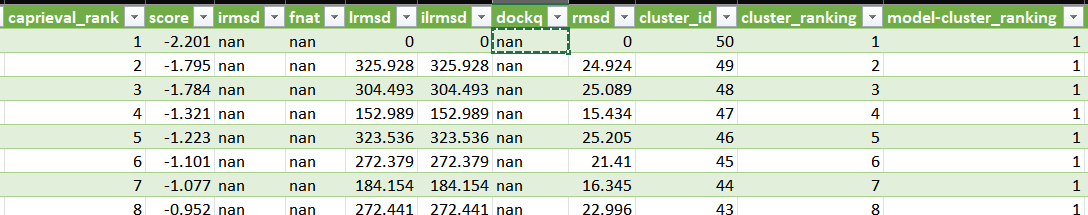
Is it sufficient to leave the next line under [caprieval] empty and unprocessed as shown in the picture?
Yes, if you do not have a reference structure.
In that case, the lowest energy structure will be used as reference for the plots
Thank you very much for your explanation, it has helped me a lot
Hii! I have a doubt! I have done the same but the output files show no scores for i-RMSD, Fnat and DOCKQ and I have got the entries as ‘nan’. Can you please help me with this?
What kind of system are you docking?
Looking at the lrmsd is looks like your molecules are not docked, i.e. no interface can be defined for the analysis.
Visualize a few PDBs to confirm
I am doing antibody antigen modelling. I did get some errors stating ‘capri WARNING] No reference contacts found’ and ‘[libparallel WARNING] Exception in task execution: list index out of range’. But I still got the scores. And when I visualised the pdb, the results were not a complex, but the Ag and Ab were present as separate chains. I did not get any other warnings too. So I am a bit unsure as where is the mistake!
Probably an issue in your restraints definition.
Did you follow first our haddock3 antibody-antigen tutorial?
https://www.bonvinlab.org/education/HADDOCK3/HADDOCK3-antibody-antigen/
Yeah I did! Thank you! I will try troubleshooting the errors
I am very happy to see that you have solved the issue of not having antigen-antibody complexes after this docking, but instead having them appear separately. I encountered a similar situation when I first started using it.
jay_14 via BioExcel <notifications@bioexcel.discoursemail.com> 于2025年9月20日周六 01:49写道:
Thank you for helping me out! It was actually an issue with the restraints definition!
I have another doubt! Where will I be able to find the Z scores after docking? I am running Haddock locally, so once I get the result files, I am not able to find the Z scores. Can you please help me with that?
Forget about the z-score. Use simply the HADDOCK score.
The caprieval modules together with the final analysis in haddock3 will generate html reports files
I have got 4 models in each cluster, so basically I have to consider the model that has the least negative HADDOCK score right? Or is there any other parameter that I should consider additionally?
The most negative score is the best - but clusters might overlap in score when you consider the standard deviation of the scores.