When NADP was docked into a coenzyme binding pocket of one enzyme using HADDOCK, in the best cluster structure, NADP molecule was split into two parts and lacks partial atom coordinates. I need some suggestions about how to deal with it.
First of all, HADDOCK only keeps the polar hydrogens. Further are you sure it is split or is it only a visualisation problem?
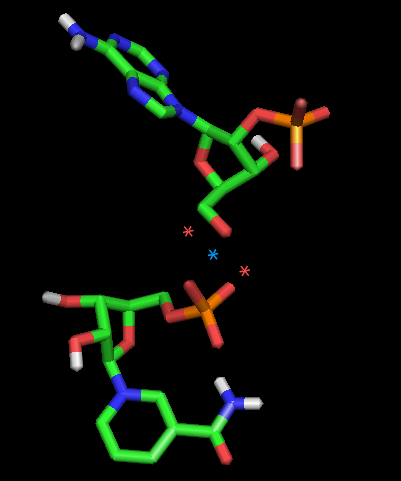
Thanks! I observed that the docked NADP visualized by Pymol yields a abnormal representation as shown in attached figure. Interestingly, ligand visualization by UCSF Chimera produces a normal structure.